The Impact of mIDH
in MDS and AML
IDH mutations are considered early “driver” mutations in the progression of MDS, as MDS-to-AML transformation occurs at a higher rate in patients with IDH1 mutations.1,2
Due to their overarching role in epigenetic regulation, there is some evidence that mutations in IDH, especially IDH1, may lead to worse outcomes in both MDS and AML.1,3–5
While meta-analyses and early data suggest an overall negative prognostic impact of IDH mutations (especially IDH1), the literature is somewhat mixed.6,7

Multiple factors can modulate the prognostic impact of IDH mutations in AML and MDS, including:7,9–12
- Isoform (IDH1 vs IDH2)
- Co-mutations
- Patient Age
- Specific Substitution / Variant
- Study Design
IDH1 vs. IDH2
Isoform may
modulate impact
of IDH mutations
MDS and AML studies have revealed important prognostic differences between patients with IDH1 and IDH2 mutations and IDH wild-type.1,4
When IDH1 and IDH2 mutant populations are combined, this may mask the prognostic value of each.7,9
Research on the impact of pooled mIDH on overall survival (OS) and event-free survival (EFS) shows mixed results.7,8,11,13,14 Some studies indicate that mIDH1 is associated with decreased survival, others suggest that mIDH2 is associated with increased survival, and certain studies only show a significant effect in certain subgroups.7,8,11,13–15
Meta analyses suggest IDH1 has a negative prognostic impact while IDH2 may have a positive or neutral prognostic impact.6,7

Grouped IDH mutations
(IDH1 and IDH2) suggest a neutral
to slight negative impact.7
Survival of AML patients with IDH mutations. Pooled HRs and 95% CI for OS (HR 1.05, P=0.5836).7

IDH1 mutations suggest
a stronger negative impact.7
Survival of AML patients with IDH1 mutations. A, Pooled HRs and 95% CI for OS (HR 1.17, P=0.0047).7

IDH2 mutations suggest
a positive impact.7
Survival of AML patients with IDH2 mutations. A, Pooled HRs and 95% CI for OS (HR 0.78, P=0.0053).7

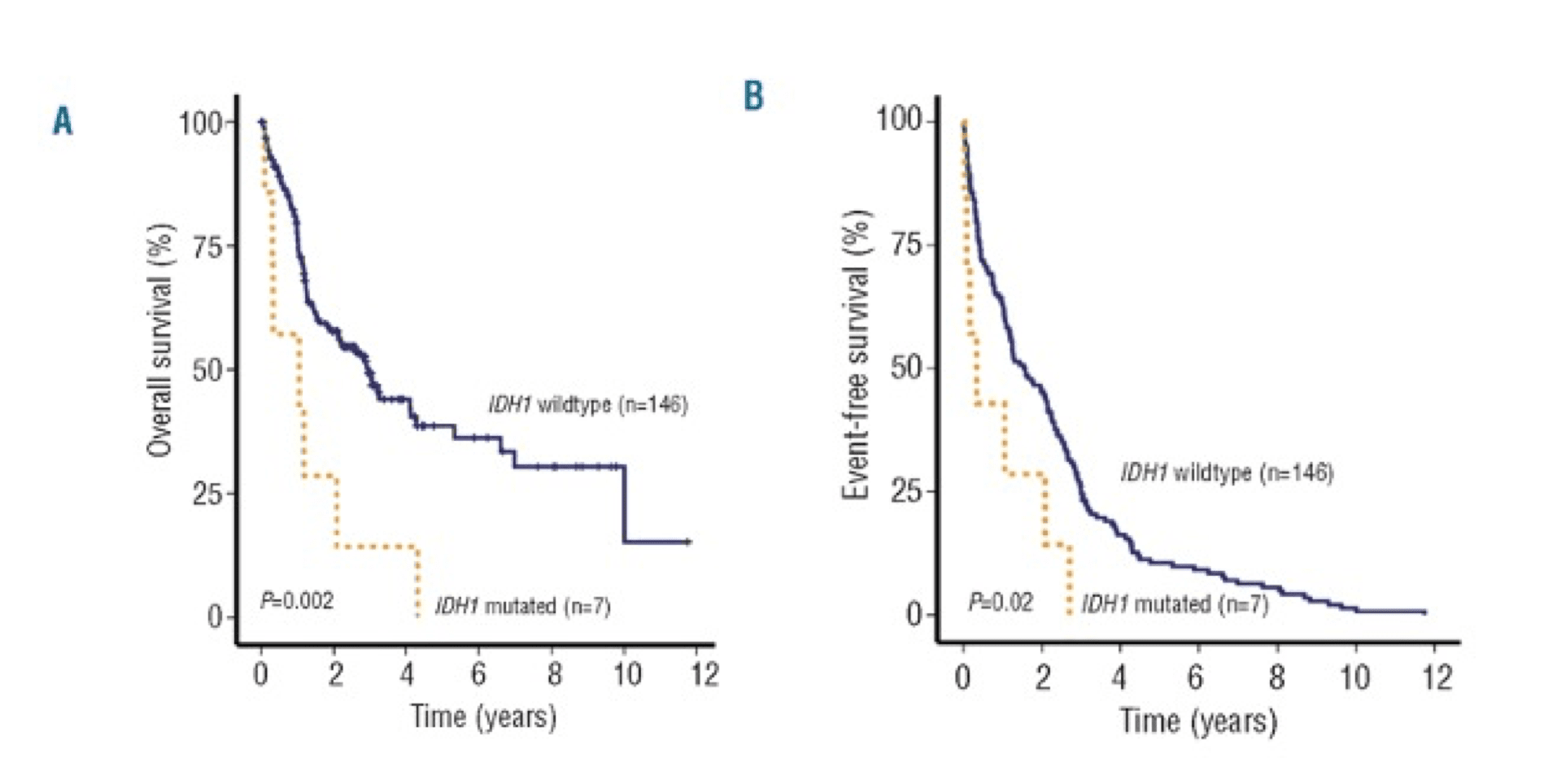
IDH1 mutations were associated with worse OS and EFS outcomes in patients with intermediate-risk NK-AML (wild-type NPM1 without FLT3-ITD).16

AML patients with mIDH1/wtNPM1 have inferior survival probability compared to patients with mIDH2/wtNPM1.10
NPM1?
DMT3A?
FLT3-ITD?
Co-mutations may
modulate the impact
of IDH mutations
Common co-mutations occurring with mIDH include NPM1, DNMT3A, and FLT3-ITD.11,12,17
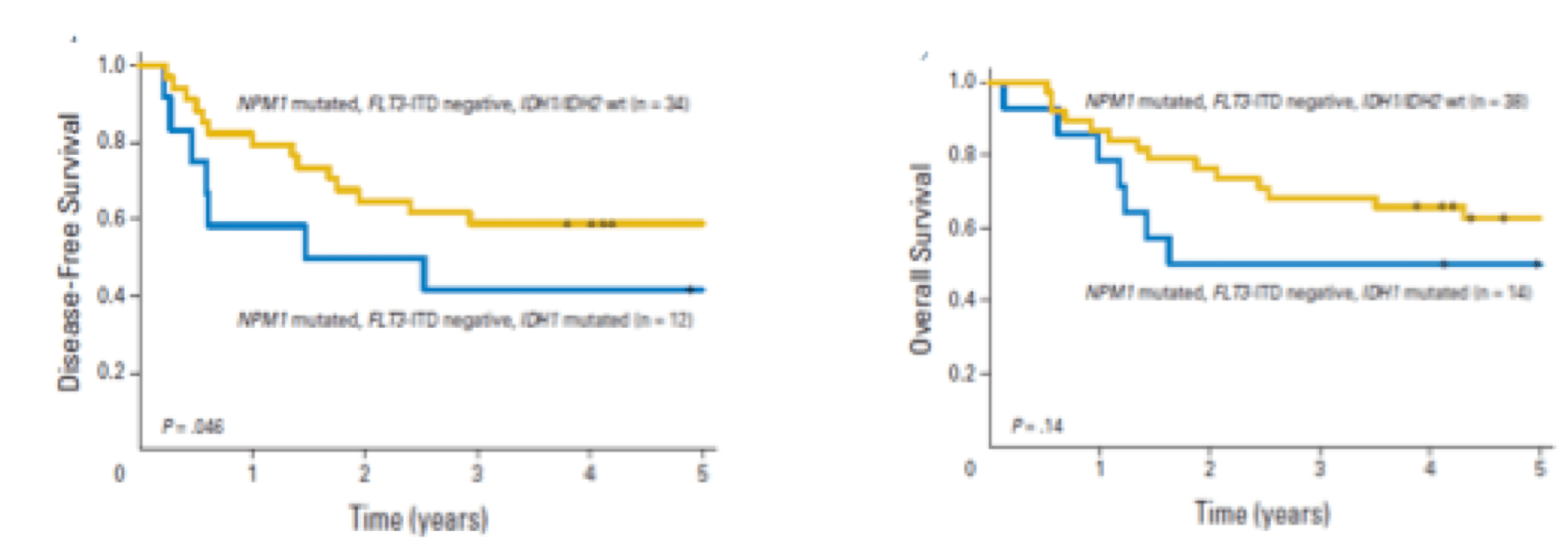
Co-mutations in DNMT3A and FLT3-ITD impart a negative prognostic impact, while NPM1 mutations impart a favorable impact.11
Patients with IDH-mutated AML without co-occurring NPM1 or with additional mutations (e.g., NPM1/DNMT3A, NPM1/FLT3-ITD) AML had especially inferior outcomes regardless of age, IDH isoform or variant.11
< 60yr
> 60yr
Patient age may
modulate impact of
IDH mutations
While the presence of an NPM1 mutation may balance out the negative impact of IDH in AML patients <60 years, older patients with mIDH had poor outcomes regardless of NPM1 status.11
IDH mutations in AML have been shown to accumulate with age, and the frequency of mIDH1 increases with the progression along the MDS-AML spectrum.11,18,19
Overall, older patients with mIDH AML have worse survival outcomes compared to younger patients.11

IDH mutations in AML accumulate with age.11
R132C
R132H
Several IDH mutational variants (location and substitution) have been studied for their impact on patient outcomes.4,12,13 In mIDH1 the R132C SNP is associated with shorter overall survival (OS) and a lower complete remission rate compared to R132H.12,13 In IDH2, the mIDH2 R172 mutation is associated with decreased OS and DFS over R140.4,12,13

Reduced OS and DFS with IDH2R172 vs IDH2R140.12

Isoform, variant, and co-mutations may influence IDH impact. OS analysis of the impact of
frequent co-mutations for IDH1/2 mutational subtypes.13
Study design may modulate
the impact of IDH mutations
Study design may modulate the impact of IDH mutations

In many studies, populations with varying age, co-mutation, isoform and variant status are combined, which reduces specificity for each cohort.7,9,13,20
Understanding how variations in study design can affect results is crucial.7 In the literature on IDH prognostic impact, significant differences between mIDH1 and mIDH2 are lost when patients with these mutations were combined into a single dataset.7 Additionally, outcomes data that address prognostic impact may also be dependent on the therapeutic paradigm at the time of data collection.12 Careful consideration of these factors helps us interpret studies of IDH mutations.12,13
Snapshot of IDH Mutation
Impact and Modifiers1,2,4,5,11–13,a
Snapshot of IDH Mutation Impact and Modifiers1,2,4,5,11–13,a

There is a trend of a negative prognostic impact for IDH mutations in MDS and AML, with the following factors associated with an especially negative impact: mIDH1, R132C, >60 years, wtNPM1, and wtDNMT3A.1,2,4,5,11–13
Visualizing Prognostic Risk in MDS
IDH1 and 2 mutations seem to have a more consistently negative prognostic impact in MDS than in AML.4
The International Working Group for Prognosis in MDS (IWG-PM), recently created a clinical-molecular prognostic model (IPSS-Molecular [IPSS-M]), to derive a unique risk score for individual MDS patients .21
Stay in the Know
Keep up to date on the latest in
IDH science from leading experts.
References:
1. Thol F, Weissinger EM, Krauter J, et al. Haematologica. 2010;95(10):1668-1674. doi:10.3324/haematol.2010.025494 2. DiNardo CD, Jabbour E, Ravandi F, et al. Leukemia. 2016;30(4):980-984. doi:10.1038/leu.2015.211 3. Cairns RA, Mak TW. Cancer Discov. 2013;3(7):730-741. doi:10.1158/2159-8290.CD-13-0083 4. Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Leukemia. 2017;31(2):272-281. doi:10.1038/leu.2016.275 5. Jin J, Hu C, Yu M, et al. PLoS ONE. 2014;9(6):e100206. doi:10.1371/journal.pone.0100206 6. Feng JH, Guo XP, Chen YY, Wang ZJ, Cheng YP, Tang YM. Am J Blood Res. 2012;2(4):254-264. 7. Xu Q, Li Y, Lv N, et al. Clin Cancer Res. 2017;23(15):4511-4522. doi:10.1158/1078-0432.CCR-16-2628 8. Marcucci G, Maharry K, Wu YZ, et al. J Clin Oncol. 2010;28(14):2348-2355. doi:10.1200/JCO.2009.27.3730 9. Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Acute Myeloid Leukemia V.3.2024. © National Comprehensive Cancer Network, Inc. 2024. All rights reserved. Accessed June 14, 2024. To view the most recent and complete version of the guideline, go online to NCCN.org.. 10. Lachowiez CA, Reville PK, Kantarjian H, et al. Am J Hematol. 2022;97(11):1443-1452. doi:10.1002/ajh.26694 11. Zarnegar-Lumley S, Alonzo T, Gerbing RB, et al. Blood Adv. Published online June 2, 2023:bloodadvances.2022008282. doi:10.1182/bloodadvances.2022008282 12. Duchmann M, Micol JB, Duployez N, et al. Blood. 2021;137(20):2827-2837. doi:10.1182/blood.2020010165 13. Middeke JM, Metzeler KH, Röllig C, et al. Blood Adv. 2022;6(5):1394-1405. doi:10.1182/bloodadvances.2021004934 14. Salem D, El-Aziz SA, El-Menshawy N, Abouzeid T, Ebrahim M. Indian J Hematol Blood Transfus. 2017;33(1):49-55. doi:10.1007/s12288-016-0649-z 15. Chou WC, Lei WC, Ko BS, et al. Leukemia. 2011;25(2):246-253. doi:10.1038/leu.2010.267 16. Abbas S, Lugthart S, Kavelaars FG, et al. Blood. 2010;116(12):2122-2126. doi:10.1182/blood-2009-11-250878 17. Messina M, Piciocchi A, Ottone T, et al. Cancers. 2022;14(12):3012. doi:10.3390/cancers14123012 18. Testa U, Castelli G, Pelosi E. Cancers. 2020;12(9):2427. doi:10.3390/cancers12092427 19. Makishima H, Yoshizato T, Yoshida K, et al. Nat Genet. 2017;49(2):204-212. doi:10.1038/ng.3742 20. Walsh C, Hunter A, Lasho T, et al. Leukemia. 2022;36(6):1693-1696. doi:10.1038/s41375-022-01551-y 21. Bernard E, Tuechler H, Greenberg PL, et al. NEJM Evid. 2022;1(7). doi:10.1056/EVIDoa2200008
NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.